Yesterday Jurrian de Kanter, PhD student of the Van Boxtel group, defended his thesis entitled: "Finding the hidden patterns: single-cell comics to reduce late effects" cum laude. We want to congratulate him with this great achievement!
Read MoreVan Boxtel Lab
"To beat cancer, we need to know how it starts."
About

This website is not an official channel of the Princess Maxima Center. To find our lab on the official platforms, click on the icons below.


Our research
Why do certain individuals develop cancer and others don’t? And why do children get cancer? Our vision is that by studying mutations in normal cells, we obtain insight into the etiology of cancer. We think this knowledge is crucial to improve cancer diagnostics and treatment, as well as for developing preventive strategies.

On the origin of cancer: studying somatic evolution in normal tissues
Identifying the rate limiting steps of cancer initiation in human tissues is challenging as many factors can play a role. The mutations in the genomes of cells can serve as an archive of their life history. We aim to decode these archives in order to pinpoint the initiation of cancer and identify causal processes in human tissues. To study the etiology of cancer, we have 3 research themes in our lab.
Learn More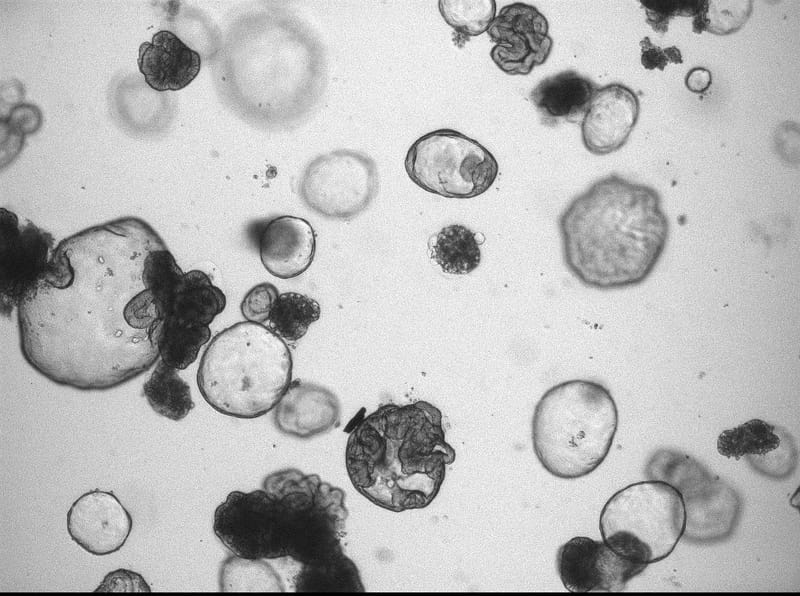
Theme 1: Tissue-specific mutation accumulation in human stem cells
Organ-specific cancer incidence varies significantly throughout the human body, which cannot be solely explained by different exposures to mutagenic environmental. Adult stem cells are likely the cellular targets for accumulation of pre-cancerous successive oncogenic hits, which eventually can give rise to tumor development, owing to their life-long capacity to propagate mutations to both self-renewing progeny and downstream progenitors. We aim to identify and study the mutational processes that are active in adult stem cells of various organs and precede oncogenic transformation.
Learn More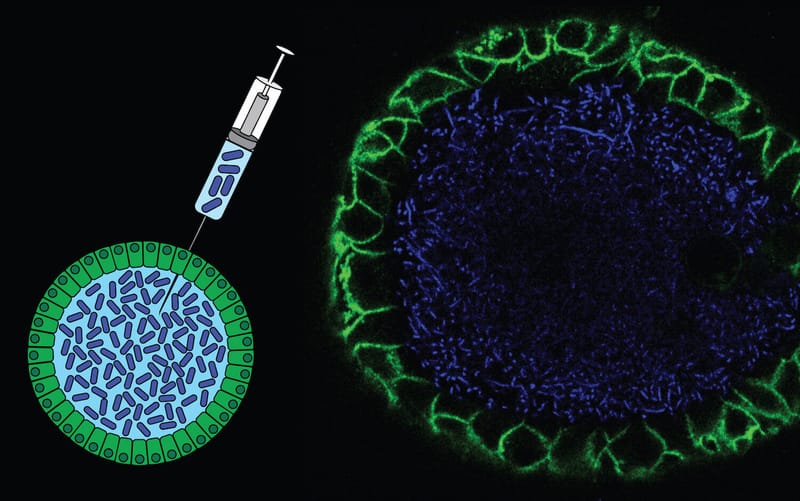
Theme 2: Tracking the origin of cancer
DNA is the largest biomolecule in the cells, which unlike other biomolecules is irreplaceable. The processes causing mutations leave characteristic patterns in the DNA, which can serve as a functional readout of mutagenic and/or DNA repair activity. In addition, phylogenetic relationships between different cells of the same individual can be exploited measure clonal dynamics within tissues. We aim to identify and study the mechanisms underlying characteristic mutation patterns in cancers as well as use mutations to retrospectively trace the cellular origin of cancer.
Learn More
Theme 3: The etiology of therapy-related malignancies in cancer survivors
Most chemotherapeutic drugs act by fatally damaging the DNA or blocking the replication thereof. However, noncancerous cells are also damaged by treatment, which can result in the accumulation of DNA mutations in normal tissues with potentially adverse effects later in life, such as novel malignancies. Our goal is to study the mutational effects of cancer treatment in normal tissues of children in order to develop novel treatment strategies aimed at minimizing or preventing adverse late effects.
Learn MoreNews
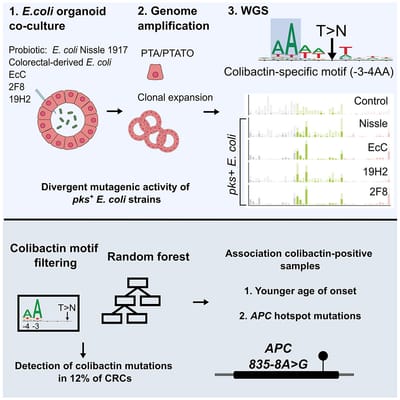
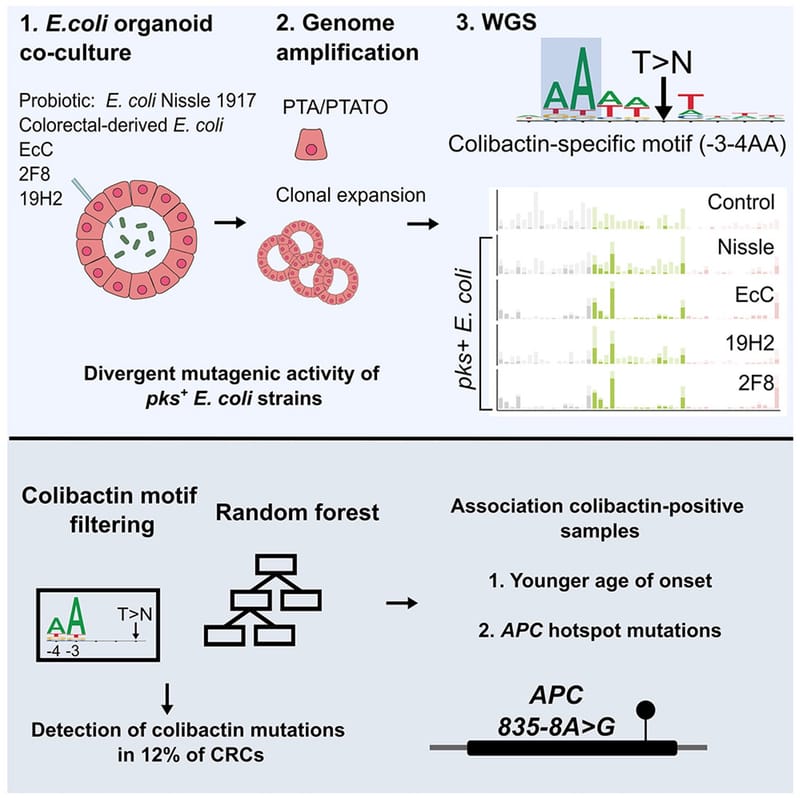
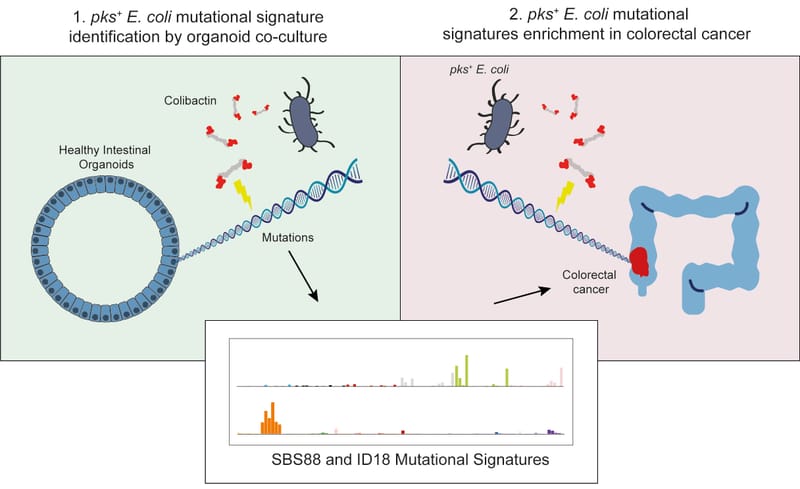
Co-culture of intestinal organoids with a colibactin-producing pks+ E. coli strain (EcC) revealed mutational signatures also found in colorectal cancer (CRC).
Read MoreRuben, has been awarded an NWO Vici grant. With the prestigious grant, Van Boxtel can further expand his research line into the late effects of childhood cancer and how these are reflected at a molecular cell level.
Read MoreJust before Christmas on December 21st, our PhD student Flavia Peci defended her thesis "Genomic Safety of Transplantation; characterising the Mutational Consequences of Treatment in Hematopoietic Stem Cells".
Read MoreTuesday our PhD student Eline Bertrums defended her thesis defence entitled: "The Clonal Dynamics underlying the genesis and regression of myeloid disorders", with cum laude.
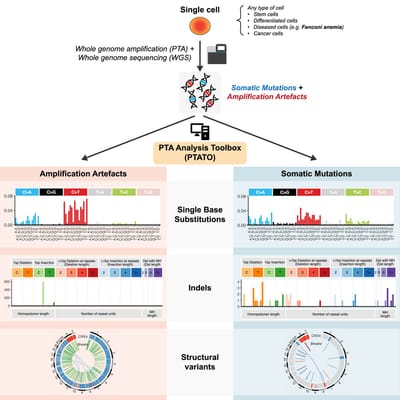
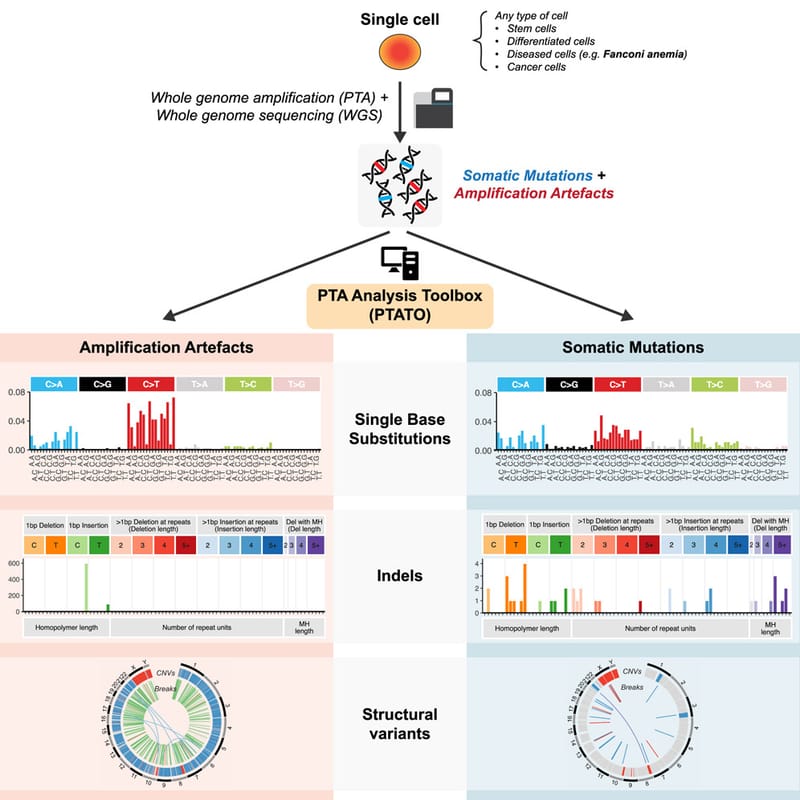
Read MoreIn our lab we developed the computational PTA Analysis Toolbox (PTATO) that can effectively filter artifacts from PTA-based WGS data, enabling accurate analyses of somatic mutations in single cells at nucleotide resolution.
Read MoreTeam
Laurianne Trabut
Technician
Matteo Boretto
Postdoc
Bo Scherer
Postdoc
Alexander Steemers
PhD student
Anais van Leeuwen
PhD student
Eirini Daskalaki
PhD student
Sam Steffens
Internship student
Catarina Leal Agostinho
Internship student
Sabine Visser
Internship student
Publications
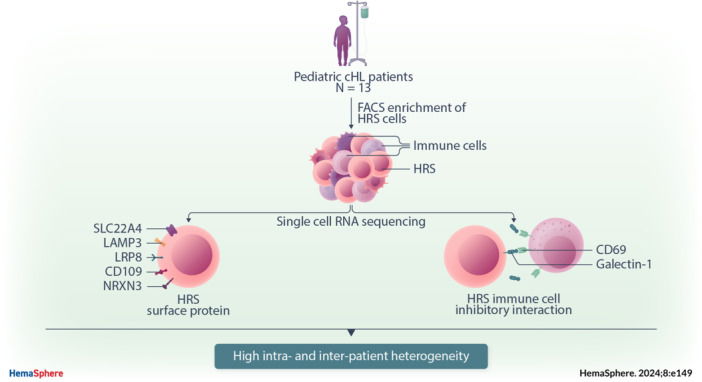
Pediatric classic Hodgkin lymphoma (cHL) patients have a high survival rate but suffer from severe long‐term side effects induced by chemo‐ and radiotherapy. This study identifies new potential therapeutic targets for cHL and highlights the importance of studying heterogeneity when identifying therapy targets, specifically those that target tumor‐immune cell interactions.
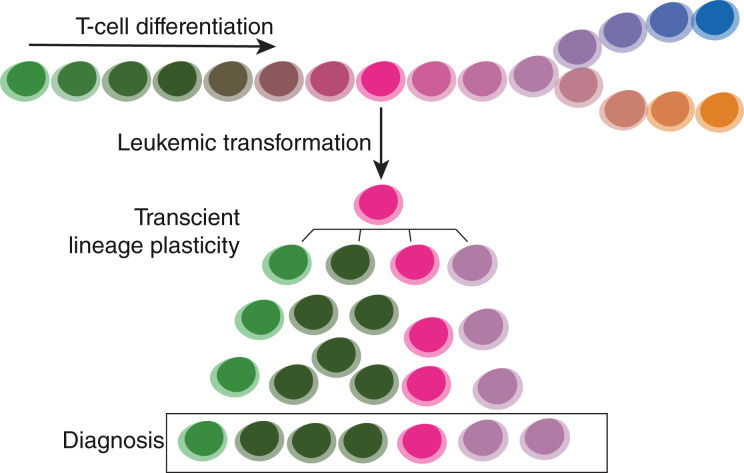
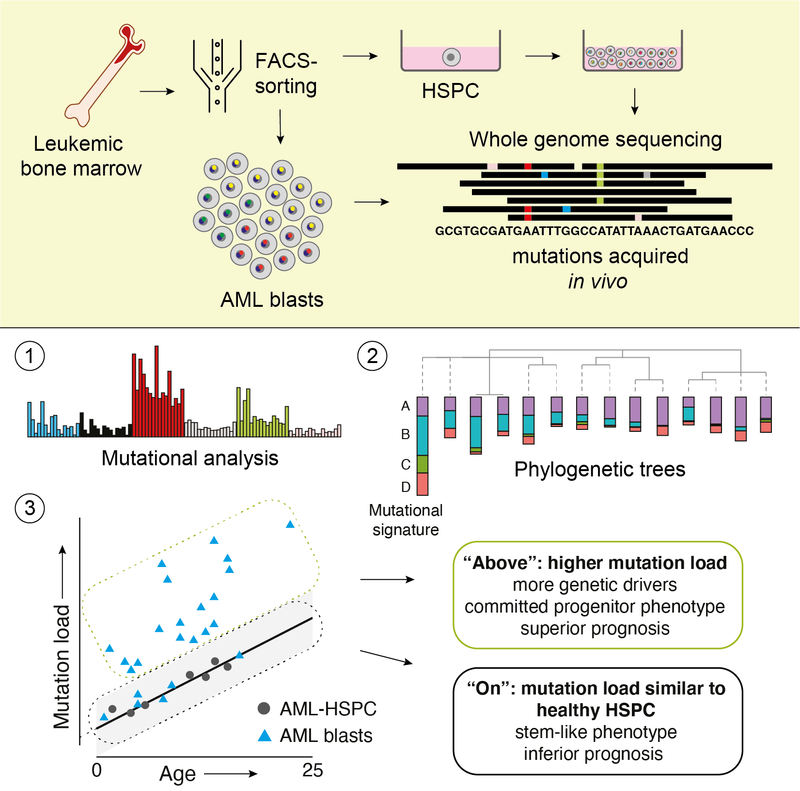
Read MoreA method merging multicolor flow cytometry with single-cell whole genome sequencing to couple cell identity with clonal lineages uncovers differentiation-state plasticity in leukemia, reconciling blocked differentiation with phenotypic plasticity in cancer.
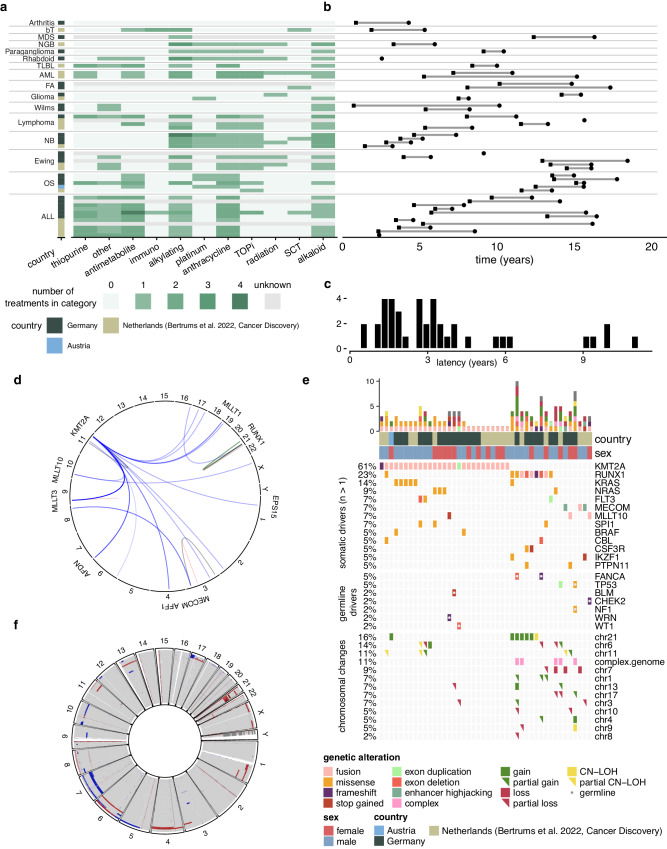
Read MoreTherapy-related myeloid neoplasms (t-MN) arise as a complication of chemo- and/or radiotherapy. Our results demonstrate that germline aberrations can interact with treatment exposures in inducing t-MN, which is important for the development of more targeted, patient-specific treatment regimens and follow-up.
Read MoreIn this study, Rosendahl Huber et al. show the mutagenic properties of pks E. coli strains, including probiotic E. coli Nissle 1917, using the extended target sequence context of colibactin and with a machine-learning model. These approaches allow for better distinguishing of colibactin-associated colorectal cancer cases, which are younger and are enriched for APC mutations matching the colibactin motif.
Read MoreDetection of somatic mutations in single cells is challenging, in part because whole-genome amplification causes many artificial mutations. Middelkamp et al. developed the computational PTA Analysis Toolbox (PTATO) that can effectively filter artifacts from PTA-based WGS data, enabling accurate analyses of somatic mutations in single cells at nucleotide resolution.
Read MoreChemotherapy increases the mutation burden of normal blood cells in cancer survivors. Only few drugs damage the DNA directly, while in most patients, chemotherapy-induced mutations are caused by processes similar to those present during normal aging. Cancer Discovery 2022
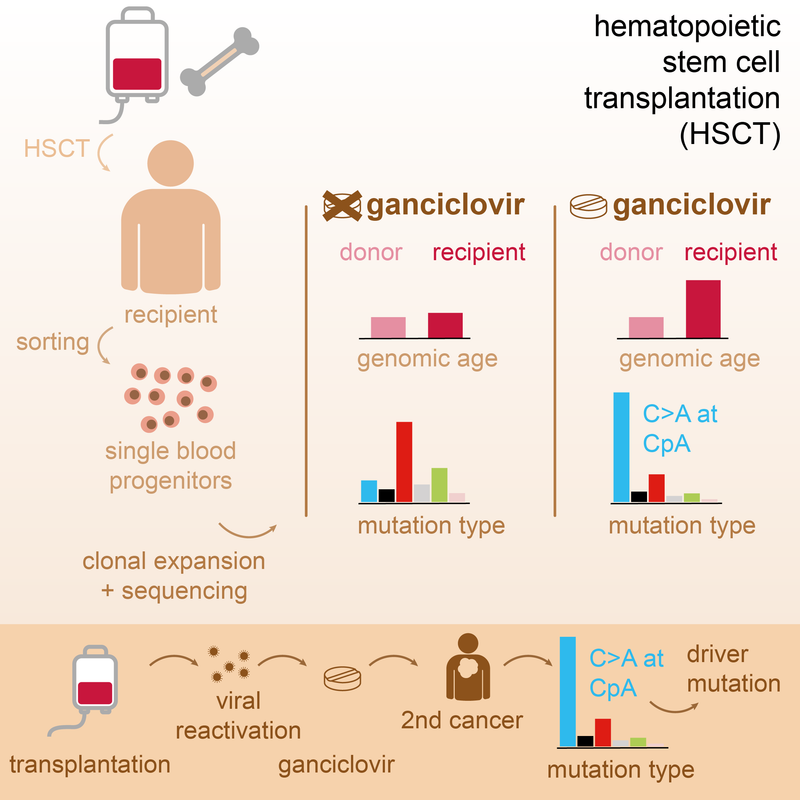
Read MoreAntiviral treatment with ganciclovir causes a unique mutational signature in stem cells of human transplant recipients. This signature was also found in therapy-related cancers and can cause cancer driver mutations. Cell Stem Cell 2021
Read MoreIn some children with acute myeloid leukemia, cancer cells have the same amount of DNA changes as healthy blood stem cells. Here, we show that these children have a poorer chance of survival compared to children whose leukemia has an above-average number of DNA changes. This study offers insight into how this form of blood cancer can develop in children. In the future, these findings may help identify which patients have a high-risk form of the disease. Blood Cancer Discovery 2021
Read MoreOur study describes a distinct mutational signature in colorectal cancer and implies that the underlying mutational process results directly from past exposure to bacteria carrying the colibactin-producing pks pathogenicity island. Nature 2020
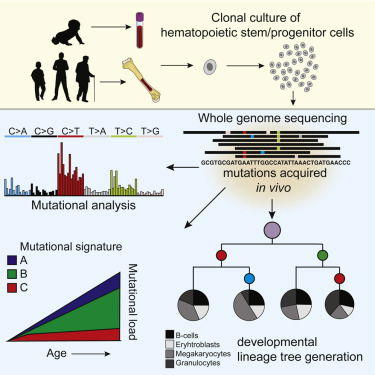
Read MoreMutation accumulation during life can contribute to hematopoietic dysfunction; however, the underlying dynamics are unknown. Using mutations found in in acute myeloid leukemia, we construct a developmental lineage tree of human hematopoiesis, revealing a polyclonal architecture and providing evidence that developmental clones exhibit multipotency. Cell Reports 2018
Read MoreHere we determine genome-wide mutation patterns in human adult stem cells of the small intestine, colon and liver of human donors with ages ranging from 3 to 87 years by sequencing clonal organoid cultures derived from primary multipotent cells. Our results show that mutations accumulate steadily over time in all of the assessed tissue types, at a rate of approximately 40 novel mutations per year, despite the large variation in cancer incidence among these tissues. Nature 2016
Read MoreLink to all publications
Read MoreERC Consolidator Grant

The outline of the project is: Therapy-related malignancies are a major cause of long-term mortality among childhood cancer survivors. However, it is unclear how exposure to chemo- and/or radiotherapy early in life induces carcinogenesis.
The aim is to determine the mechanisms and rate-limiting steps underlying the genesis of second malignancies in childhood cancer survivors. For this, we will focus on studying the etiology of therapy-related myeloid malignancies (t-MNs).
Dr. van Boxtel has pioneered methods to characterize mutation accumulation in single stem cells and study clonal lineages in the human hematopoietic system. The van Boxtel lab is embedded in Europe’s largest childhood cancer center, providing the opportunity to apply our techniques to unique patient material.
In Objective 1, we will dissect the life history of t-MN and study its cellular origin. Our key question is: Was the original t-MN clone already present before chemotherapy exposure, or generated as a consequence thereof? We will address this by tracking back clonal lineages in the hematopoietic tissue of patients using the mutations present in their second cancers.
In Objective 2, we will study the mutational consequences of chemotherapy in normal hematopoietic cells of children before and after they received treatment. Our key question is: Is enhanced mutagenesis rate limiting for t-MN development? To address this, we will perform in-depth mutational analyses and in vitro validations.
In Objective 3, we will determine phenotypic effects of chemotherapy on population dynamics of blood. Our key question is: how does chemotherapy affect selection dynamics and clonal composition of blood? To address this, we will integrate clonal histories and lineage contributions using somatically acquired mutations. Our unique methodology and anticipated novel insights will not contribute to improved survival of children with cancer, but also to increased fundamental knowledge on the origin of cancer.
Links

http://bioconductor.org/packages/release/bioc/html/MutationalPatterns.html
Github page:
https://github.com/ToolsVanBox
Alumni
Jose Ubels
Postdoc
Niels Groenen
Technician
Jurrian de Kanter
PhD Student
Flavia Peci
PhD student
Eline Bertrums
PhD student
Sjors Middelkamp
Postdoc
Inge van der Werf
Postdoc
Freek Manders
PhD Student
Karlijn Hasaart
PhD Student
Axel Rosendahl Huber
PhD student
Arianne Brandsma
Postdoc
Rurika Oka
Bioinformatician